
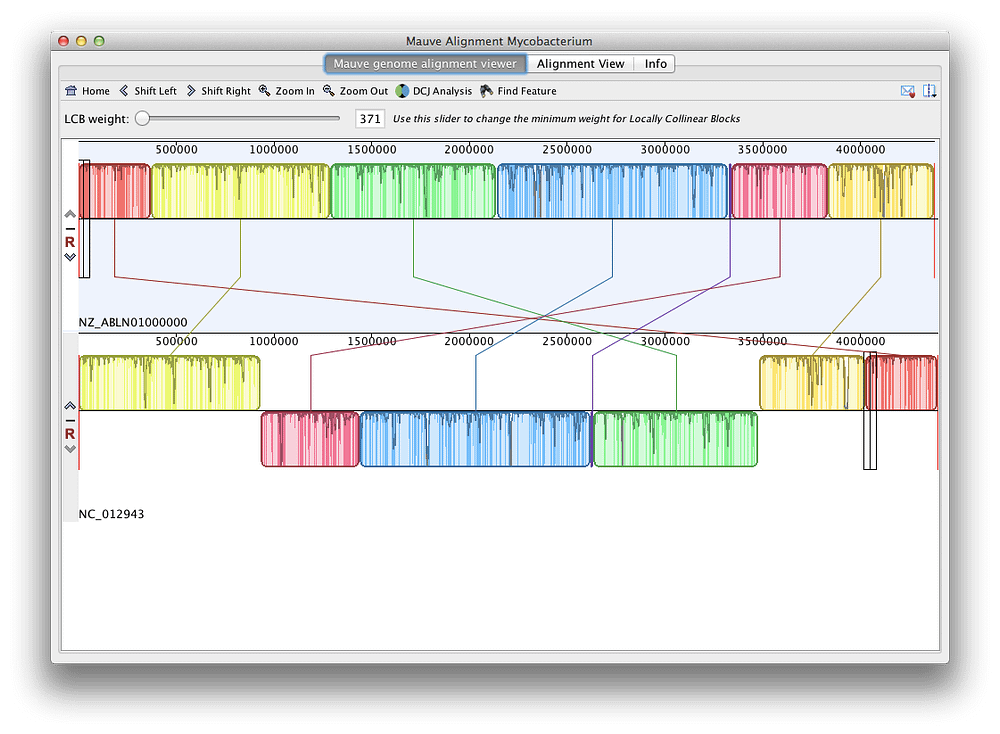
A maximum likelihood tree was inferred by employing the LG + G + I + F model of amino acid substitution (number of substitution rate categories, 4 gamma shape parameter, 0.796 proportion of invariable sites, 0.222) in the PhyML 3.0 Web server ( 13). Amino acid sequences of lef-8 and pif-2 gene products were aligned with MUSCLE ( 11) using the default settings and concatenated after removing ambiguously aligned residues using the -gappyout command in trimAl version 1.2 ( 12). Although the closest relationship identified was between ChdiNPV and ChmuNPV, several divergent regions were detected in an alignment conducted using progressiveMauve ( 10) implemented in Geneious Prime 2019.2.3 (Biomatters) using the default settings ( Fig. 1B), which contributed to the discrepancy of the presence or absence of ORFs in each viral genome.įIG 1 FIG 1 (A) Phylogenetic tree of alphabaculovirus. ChroNPV was aligned to this lineage ( Fig. 1A). Phylogenic analysis using concatenated sequences of LEF-8 and PIF-2 indicates that ChdiNPV belongs to the group I alphabaculoviruses and is the closest to ChmuNPV in lineage, including ChocNPV and CfMNPV.

The mean amino acid identities of ChdiNPV putative proteins with orthologous proteins of the above-mentioned 4 viruses were 98.3%, 87.7%, 87.4%, and 84.4%, respectively. A blastp search with a cutoff E value of 1 × 10 −5 using blast+ ( 9) found that ChdiNPV shared 140, 136, 136, and 136 ORFs with ChmuNPV, ChocNPV, CfMNPV, and ChroNPV, respectively. Global alignment using EMBOSS Stretcher ( 8) showed that the ChdiNPV genome had 95.4%, 81.6%, 81.2%, and 75.8% nucleotide identities to ChmuNPV, ChocNPV, CfMNPV, and ChroNPV, respectively. The remaining 144 ORFs had similarities to other baculovirus genes, including all baculovirus core genes.

diversana NPV (ChdiNPV) was 122,827 bp long with a G+C content of 50.2% and was estimated to have 150 ORFs, including 6 ORFs unique to ChdiNPV. Putative open reading frames (ORFs) encoding more than 50 amino acids were identified using the Glimmer 3 version 1.5 ( 7) algorithm trained for ChmuNPV ORF annotations using Geneious Prime 2019.2.3 (Biomatters) and were manually edited. The genome sequence reported here used the sequence of the longer amplicon. The PCR generated two amplicons (482 and 656 bp) containing repeat sequences. An ambiguous region with lower coverage inside the orf1629 gene was amplified using PCR, and the DNA was sequenced using Sanger sequencing. All processed reads were then used to assemble the circular contig, permitting 20,940,902 reads to be assembled and resulting in a mean coverage of 16,856× (standard deviation, 2,051×). This generated a contig with overlapping sequences at both ends, from which a circular contig was created. Assembly was conducted first using 5% of the total processed reads (1,052,696 of 21,053,934 reads) using de novo assembly in Geneious Prime 2019.2.3 (Biomatters) with default parameters of low sensitivity/fastest setting except that the “only use paired hits during assembly” option was used. Trimming of low-quality ends (Phred quality scores of
diversana collected at a Todomatsu fir stand in Hokkaido, Japan, which had been preserved at the Forestry and Forest Products Research Institute since 1972. As per a previously described method ( 6), virions were purified from viral occlusion bodies prepared from three larval samples of C.


 0 kommentar(er)
0 kommentar(er)
